5.7 Drawing secondary structures
![]()
![]()
![]() Next: 6. Notes
Up: 5. Methods 3
Previous: 5.6 Folding
at
Next: 6. Notes
Up: 5. Methods 3
Previous: 5.6 Folding
at
5.7 Drawing secondary structures
The GCG package [22] has a number of programs (SQUIGGLES, CIRCLES, DOMES, MOUNTAINS AND DOTPLOT) that produce graphical representations of RNA secondary structures. The input to these programs is a ``CONNECT'' file that is like a ct file, except for the first two lines. Thus ct files have to be edited to be used by these GCG programs. There is a small file ct_gcg.f that is distributed with the mfold package that contains a subroutine for producing GCG format ct files. This subroutine can be used to replace the existing ct subroutine in the mrna.f file (mrna1.f in the X11 versions).
The ``draw'' program by Shapiro et al. [31]
accepts region file and sequence input. This program is highly interactive, and was
used to produce Figure 5. It has recently been ported to the Silicon Graphics IRIS
platform, where it makes use of the DISSPLA![]() package
for producing output files. The ``naview'' program by Bruccoleri and Heinrich [32]accepts ct file input and also produces a pleasing output.
It has been adapted to run in a general UNIX environment. The output is a device
independent plot that requires the PLT2 plotting package [33] to produce output.
package
for producing output files. The ``naview'' program by Bruccoleri and Heinrich [32]accepts ct file input and also produces a pleasing output.
It has been adapted to run in a general UNIX environment. The output is a device
independent plot that requires the PLT2 plotting package [33] to produce output.
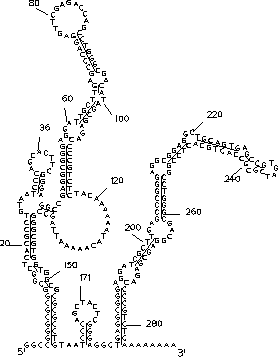
Figure 5: Output of the ``draw'' program (5.7) for the optimal folding of the Alu sequence from Figure 1.
Michael Zuker
Thu Nov 2 14:28:14 CST 1995