AUXILIARY AND INDIVIDUAL PROGRAMS
Next: Sample foldings Up: Running the programs Previous: Optimal and suboptimal foldings
AUXILIARY AND INDIVIDUAL PROGRAMS
The mfold package contains a script, also named mfold, that performs a folding according to information entered on the command line. This script is itself composed of scripts and (Fortran or C) executable programs, many of which can be run separately. Some of these programs are now described.
auxgen: This program creates the 'FILE_NAME.PNT' file.boxplot_ng: This program creates an energy dot plot in PostScript or gif form from 'FILE_NAME.PLOT'. The file boxplot_ng.txt contains instructions.ct_boxplot: This program creates a dot plot containing only those base pairs found in a collection of structures (in ``ct format'') that are specified on the command line. These must all be foldings of the same sequence fragment. At present, the mfold script does not use this program.ct_compare: This program compares 2 ``ct files''. The first contains a single reference structure. The second contains 1 or more foldings of the same sequence. The number of bases and helices from the first structure that are conserved in the other structures are computed and displayed. For the purposes of this program, a ``helix'' may contain bulge or interior loops of size 1 or 2 and must have at least 3 base pairs [17,40].efn: This program computes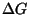 for all the foldings in a ``ct file''.
The energy rules correspond exactly to what is used in mfold.
for all the foldings in a ``ct file''.
The energy rules correspond exactly to what is used in mfold.efn2: This program computes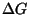 for all the foldings in a ``ct file''
using a more precise free energy computation that takes into account coaxial stacking
and Jacobson-Stockmeyer theory for multi-branched loops. (See equation 7 and Figure 8,
respectively.)
for all the foldings in a ``ct file''
using a more precise free energy computation that takes into account coaxial stacking
and Jacobson-Stockmeyer theory for multi-branched loops. (See equation 7 and Figure 8,
respectively.)h-num: This program computes an ``h-num'' file from a ``plot'' file.nafold: This is the principle folding program of the mfold package, and corresponds to the program ``lrna'' and ``crna'' in earlier versions of mfold. It has 2 command line arguments. The first takes on the values `l' or `c', for linear (default) or circular folding, respectively. The second has values ``text'' (default) or ``html'' for plain text output, and for some html output, as described above. It is run twice by the mfold script. It may be run alone, as it prompts the user for input on an interactive basis. In this mode, the user may fold all the sequences contained in a single file, an option not available with the mfold script. The instructions for running this program have been described in [19] and also on the WWW at http://mfold.bioinfo.rpi.edu/. The interactive energy dot plot is not functional in version 3.0.naview: This is a modified version of the naview program described in [41]. Actually, in mfold, naview is a script that runs a binary called naview.exe. Both naview and naview.exe may be run alone in interactive form. The mfold script uses the files ``bases.nav'' and ``lines.nav'', stored in MFOLDLIB to direct ``naview''. The first produces an output that displays individual residues as letters, while the second gives a structure outline only.newtemp: The newtemp program creates free energy files for folding RNA or DNA at different temperatures. The latest RNA parameters consist only of free energies measured at 37°. Since there are no corresponding enthalpy files, it is not possible to fold RNA at temperatures other than 37°. If the user wishes to fold RNA at different temperatures, then the ``dg'' and ``dh'' files from mfold version 2.3 should be copied into MFOLDLIB. These older free energy parameters will be supplied with mfold version 3.0. DNA folding may be done at arbitrary temperatures between 0 and 100 degrees. However, it should be remembered that the DNA loop parameters are all estimated from values published in the literature or by comparison with RNA.plt22ps: This program takes a ``plt2'' file from naview and creates a PostScript file of a plotted structure. It can use an ``ann'' file or an ``ss-count'' file to annotate with P-num or ss-count values, respectively.plt22gif: This program is the same as plt22ps, except that the structure output file is in gif format.sav2p-num: This interactive program uses an existing `fold_name.sav' file to create a P-num file.sav2plot: This interactive program uses an existing `fold_name.sav' file to create a ``plot'' file. The resulting ``plot'' file may be sorted and filtered using the command: FILTER-SORT NAME.PLOT N, where name.plot is a ``plot'' file, and N is the minimum helix size that is desired. Helices shorter than N and not removed in optimal foldings.scorer: This program is similar to ct_compare. It compares foldings helix by helix, displaying a more detailed output.split_ct.awk: This is a Unix awk script that splits a ct file into multiple files, containing 1 folding in each. These individual files may be processed by naview, plt22ps and plt22gif.ss-count: This program computes single stranded sample statistics from a collection of foldings stored in a single ct file.
Next: Sample foldings Up: Running the programs Previous: Optimal and suboptimal foldings
 |
Michael Zuker Center for Computational Biology Washington University in St. Louis 1998-12-05 |