Constrained folding
Next: Running the programs Up: Algorithms and Thermodynamics for Previous: Loops and Nearest neighbor
Constrained folding
In addition to the free energy rules, specific constraints may be used to force or prohibit base pairs. A special file containing commands to constrain folding is used. The command syntax is rigid. The various commands and syntax are given below.
- Forcing a string of consecutive bases to pair.
Syntax: F i 0 k
Ribonucleotides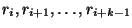 are forced to be double stranded. The partners
for these bases are chosen by the program. As an example, the command:
are forced to be double stranded. The partners
for these bases are chosen by the program. As an example, the command:
F 23 0 5
would cause bases 23, 24, 25, 26 and 27 to pair. - Forcing a string of consecutive base pairs.
Syntax: F i j k
Base pairs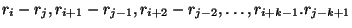 are
forced to occur. This is the same thing as forcing a helix to form. The helix is
designated by its (external) closing base pair, i.j. As an example, the
command:
are
forced to occur. This is the same thing as forcing a helix to form. The helix is
designated by its (external) closing base pair, i.j. As an example, the
command:
F 2 110 3
would force base pairs 2.110, 3.109 and 4.108. Note that these base pairs must be able to form! Be aware also that mfold filters out isolated base pairs. - Prohibiting a string of consecutive bases from pairing.
Syntax: P i 0 k
Ribonucleotides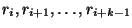 are prevented from pairing.
are prevented from pairing. - Prohibiting a string of consecutive base pairs
Syntax: P i j k
Base pairs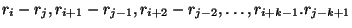 are
not allowed to form. This is equivalent to prohibiting a helix.
are
not allowed to form. This is equivalent to prohibiting a helix. - Prohibiting 1 segment of a sequence from pairing with another
Syntax: P i-j k-l
where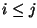 and
and 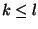 . In this case, no base pairs are allowed between
. In this case, no base pairs are allowed between
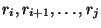 and
and
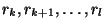 . Note that the 2 segments need not be distinct.
For example, the command:
. Note that the 2 segments need not be distinct.
For example, the command:
P i-j i-j
will not allow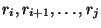 to pair with itself.
to pair with itself. - Annotated bases.
- mfold recognizes A, C, G, U and T. In RNA folding, a `T' will be treated as a `U'; and vice versa for DNA folding. In addition, B, D, H and V are recognized as A, C, G and U/T, respectively. Bases marked in this way are regarded as susceptible to nuclease cleavage. They are allowed to pair only if their 3' neighbor is unpaired. This is an old feature of mfold .
-
mfold also recognizes W, X, Y and Z as A, C, G and U/T, respectively.
These bases are regarded as ``modified'' and are allowed to pair only at the
ends of helices. At this time, the commonly used ambiguous codes shown in Table
2 are not supported by mfold .
Table 2: Unsupported ambiguous codes for RNA/DNA. mfold does not currently support the convention for ambiguous codes. Unrecognized bases will not be allowed to pair. Ambiguity A,G C,U/T A,U/T C,G A,C G,U/T Code letter R Y W S M K Ambiguity C,G,U/T A,G,U/T A,C,U/T A,C,G A,C,G,U/T Code letter B D H V N
![]()
![]()
![]()
![]()
Next: Running the
programs Up: Algorithms and Thermodynamics for Previous: Loops and Nearest neighbor
 |
Michael Zuker Center for Computational Biology Washington University in St. Louis 1998-12-05 |