Example 2
Next: EXAMPLE 3 Up: Sample foldings Previous: EXAMPLE 1
EXAMPLE 2
Important alternative foldings might not appear in the energy dot plot if
ΔΔG is too small. This is especially true in the folding of short
sequences. When the short sequence:
AAGGGGUUGG UCGCCUCGAC UAAGCGGCUU GGAAUUCC,
is folded, also with default parameters, a single optimal folding is computed. However,
the energy dot plot contains only the optimal, black dots from Figure 15. Changing the window size would not reveal anything new. When
the value of P is increased to 25 (25%), the energy dot plot now reveals a very
distinct alternate folding as shown in Figure 15. The mfold program now computes 2 foldings, plotted in Figure 16, using the default value of W.
| Figure 15: The energy dot plot for ``Example 2'' sequence ΔΔG increased to 25% of 10.1, or 2.5 kcal/mol. The value of ΔΔG in the plot may be less than this maximum value, since there may be no base pairs in foldings that are exactly ΔΔG from the minimum free energy. The 2 green dots represent base pairs that can be in foldings with ΔG between -9.4 and -8.6 kcal/mol. These numbers are -8.6 and -7.9 for the yellow dots. In this case, the black dots comprise the optimal folding, and the yellow dots comprise the single suboptimal folding that is computed. The green dots would only be found in a folding if the value of W were lowered sufficiently. | 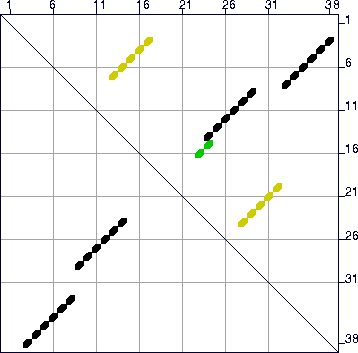 |
| Figure 16: The 2 predicted foldings for the ``Example 2'' sequence. (a) The optimal folding with ΔG = -10.1 kcal/mol. (b) The suboptimal fold (ΔG = -7.9 kcal/mol) found after refolding with `P'=25. | 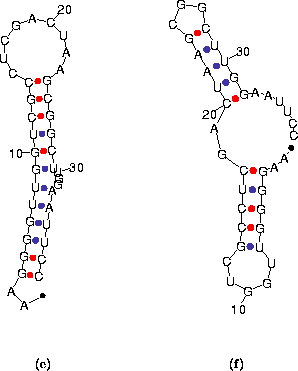 |
![]()
![]()
![]()
![]()
Next: EXAMPLE 3
Up: Sample
foldings Previous: EXAMPLE 1
 |
Michael Zuker Center for Computational Biology Washington University in St. Louis 1998-12-05 |